Simulate with all variabilities
Let’s use a simple 1-compartment model with absorption compartment to illustrate the different levels of variabilities.
refModel <- model_suite$nonmem$advan2_trans2
refModel## [MAIN]
## KA=THETA_KA*exp(ETA_KA)
## CL=THETA_CL*exp(ETA_CL)
## V=THETA_V*exp(ETA_V)
## S2=V
##
## [ODE]
## d/dt(A_DEPOT)=-KA*A_DEPOT
## d/dt(A_CENTRAL)=-CL*A_CENTRAL/V + KA*A_DEPOT
## d/dt(A_OUTPUT)=CL*A_CENTRAL/V
## F=A_CENTRAL/S2
##
## [ERROR]
## CONC=F
## CONC_ERR=CONC*(EPS_PROP + 1)
##
##
## THETA's:
## name index value fix
## 1 KA 1 1 FALSE
## 2 CL 2 5 FALSE
## 3 V 3 80 FALSE
## OMEGA's:
## name index index2 value fix type
## 1 KA 1 1 0.025 FALSE var
## 2 CL 2 2 0.025 FALSE var
## 3 V 3 3 0.025 FALSE var
## SIGMA's:
## name index index2 value fix type
## 1 PROP 1 1 0.025 FALSE var
## No variance-covariance matrix
##
## Compartments:
## A_DEPOT (CMT=1)
## A_CENTRAL (CMT=2)
## A_OUTPUT (CMT=3)We’re going to use a very basic dataset. 1000 mg QD shall be administered for a week.
ds <- Dataset(25) %>%
add(Bolus(time=0, amount=1000, ii=24, addl=6)) %>%
add(Observations(times=seq(0,24*7,by=4)))All subjects are different due to IIV and RUV.
results <- refModel %>% simulate(dataset=ds, seed=1)
spaghettiPlot(results, "CONC_ERR")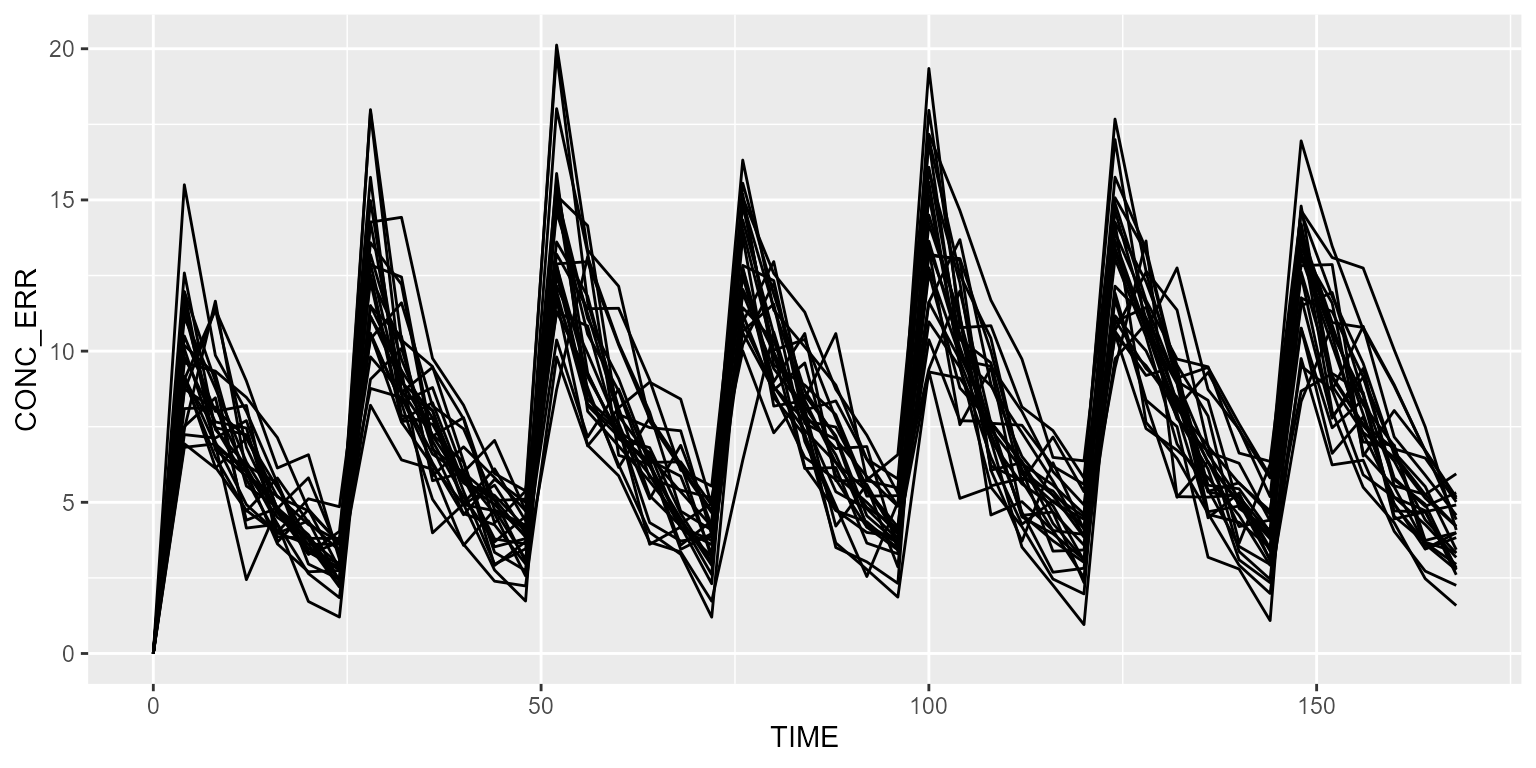
Simulate without RUV
Disabling RUV is done as follows:
## THETA's:
## name index value fix
## 1 KA 1 1 FALSE
## 2 CL 2 5 FALSE
## 3 V 3 80 FALSE
## OMEGA's:
## name index index2 value fix type
## 1 KA 1 1 0.025 FALSE var
## 2 CL 2 2 0.025 FALSE var
## 3 V 3 3 0.025 FALSE var
## SIGMA's:
## name index index2 value fix type
## 1 PROP 1 1 0 FALSE var
## No variance-covariance matrixIn that case, CONC_ERR (the observed concentration) is identical as CONC (the model-simulated plasma concentration).
results <- model %>% simulate(dataset=ds, seed=1)
spaghettiPlot(results, "CONC_ERR")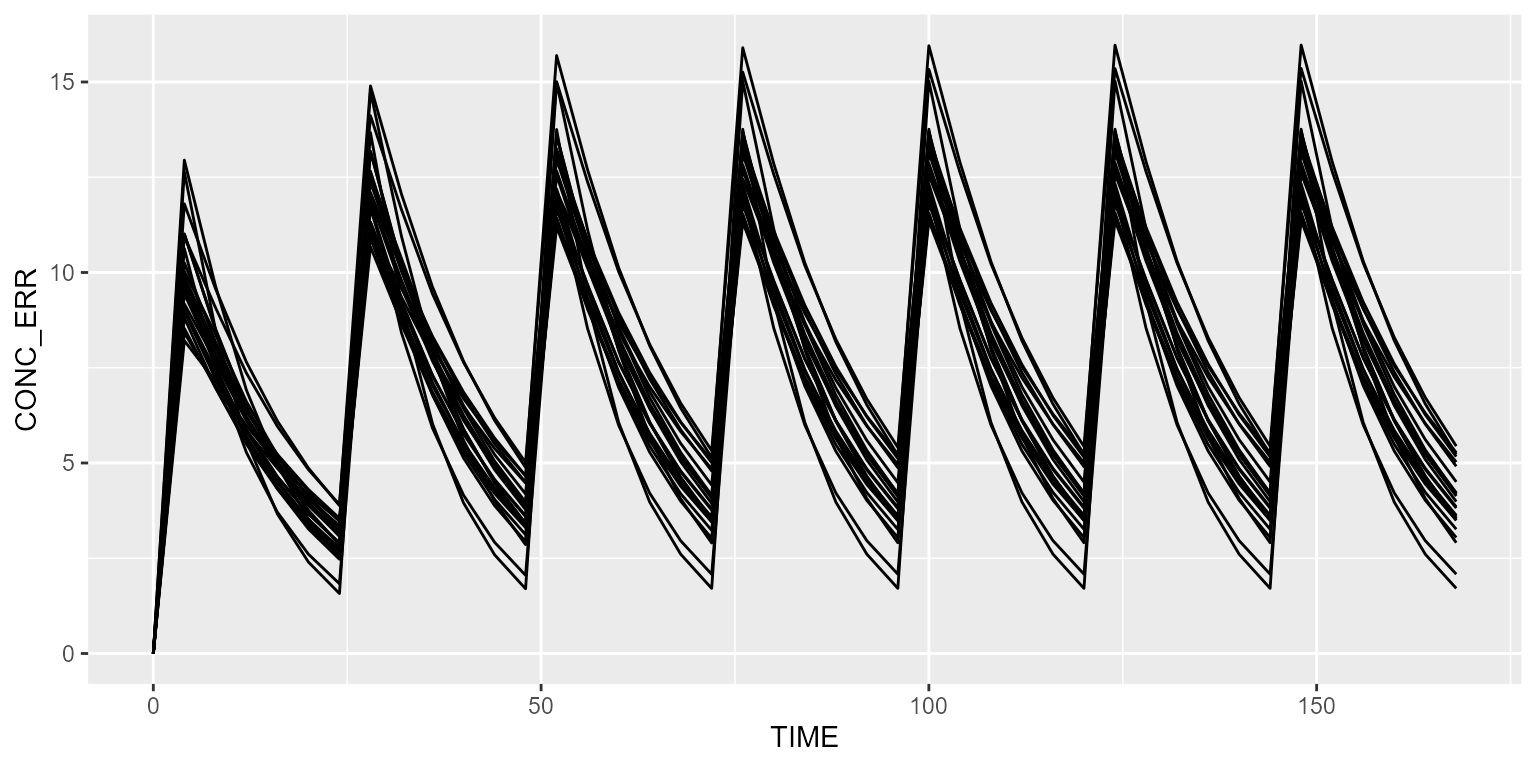
spaghettiPlot(results, "CONC")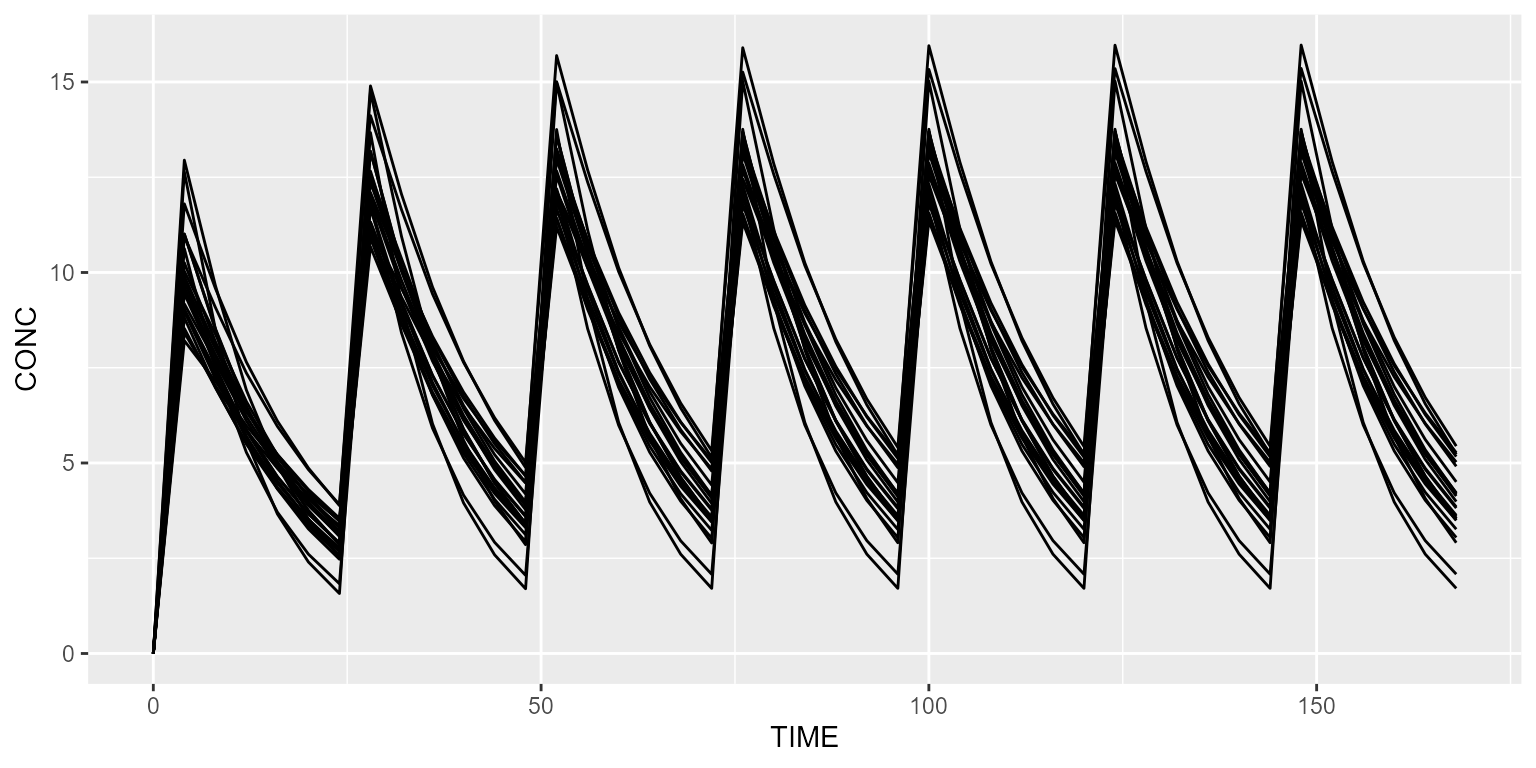
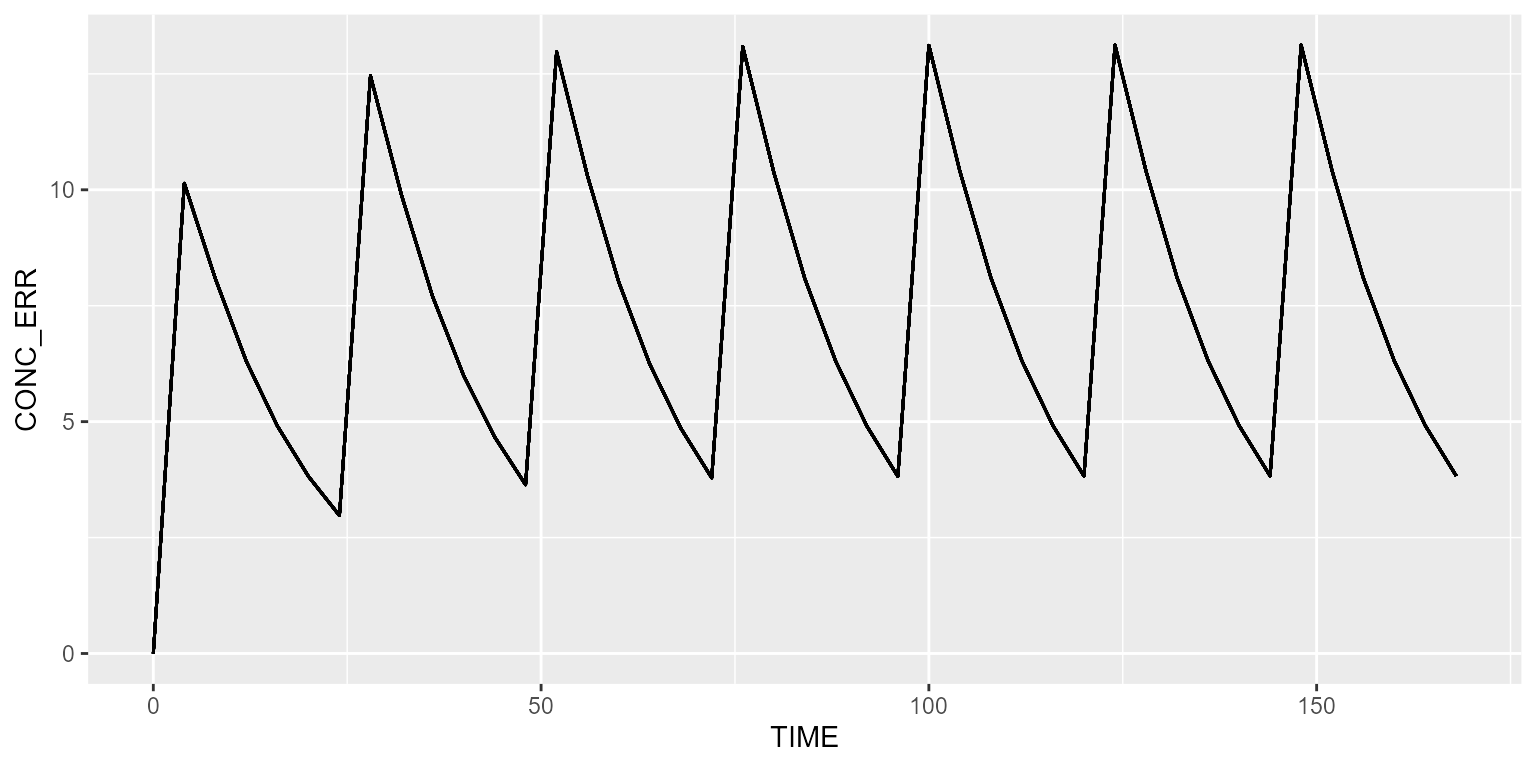
Simulate without RUV and IIV
Disabling RUV and IIV is done as follows:
## THETA's:
## name index value fix
## 1 KA 1 1 FALSE
## 2 CL 2 5 FALSE
## 3 V 3 80 FALSE
## OMEGA's:
## name index index2 value fix type
## 1 KA 1 1 0 FALSE var
## 2 CL 2 2 0 FALSE var
## 3 V 3 3 0 FALSE var
## SIGMA's:
## name index index2 value fix type
## 1 PROP 1 1 0 FALSE var
## No variance-covariance matrixNow, the typical profile is shown for all subjects.
results <- model %>% simulate(dataset=ds, seed=1)
spaghettiPlot(results, "CONC_ERR")