This vignette shows how initial conditions may be used in CAMPSIS.
Init central compartment
Assume the following 2-compartment model is used.
model <- model_suite$nonmem$advan3_trans4We’d like to init the central compartment to a predefined value,
e.g. 1000.
This can be achieved as follows.
model <- model %>% add(InitialCondition(compartment=1, rhs="1000"))The resulting model has now a new block [INIT] which
describes the initial condition:
model## [MAIN]
## CL=THETA_CL*exp(ETA_CL)
## V1=THETA_V1*exp(ETA_V1)
## V2=THETA_V2*exp(ETA_V2)
## Q=THETA_Q*exp(ETA_Q)
## S1=V1
##
## [ODE]
## d/dt(A_CENTRAL)=Q*A_PERIPHERAL/V2 + (-CL/V1 - Q/V1)*A_CENTRAL
## d/dt(A_PERIPHERAL)=-Q*A_PERIPHERAL/V2 + Q*A_CENTRAL/V1
## d/dt(A_OUTPUT)=CL*A_CENTRAL/V1
## F=A_CENTRAL/S1
##
## [INIT]
## A_CENTRAL=1000
##
## [ERROR]
## CONC=F
## CONC_ERR=CONC*(EPS_PROP + 1)
##
##
## THETA's:
## name index value fix
## 1 CL 1 5 FALSE
## 2 V1 2 80 FALSE
## 3 V2 3 20 FALSE
## 4 Q 4 4 FALSE
## OMEGA's:
## name index index2 value fix type
## 1 CL 1 1 0.025 FALSE var
## 2 V1 2 2 0.025 FALSE var
## 3 V2 3 3 0.025 FALSE var
## 4 Q 4 4 0.025 FALSE var
## SIGMA's:
## name index index2 value fix type
## 1 PROP 1 1 0.025 FALSE var
## No variance-covariance matrix
##
## Compartments:
## A_CENTRAL (CMT=1)
## A_PERIPHERAL (CMT=2)
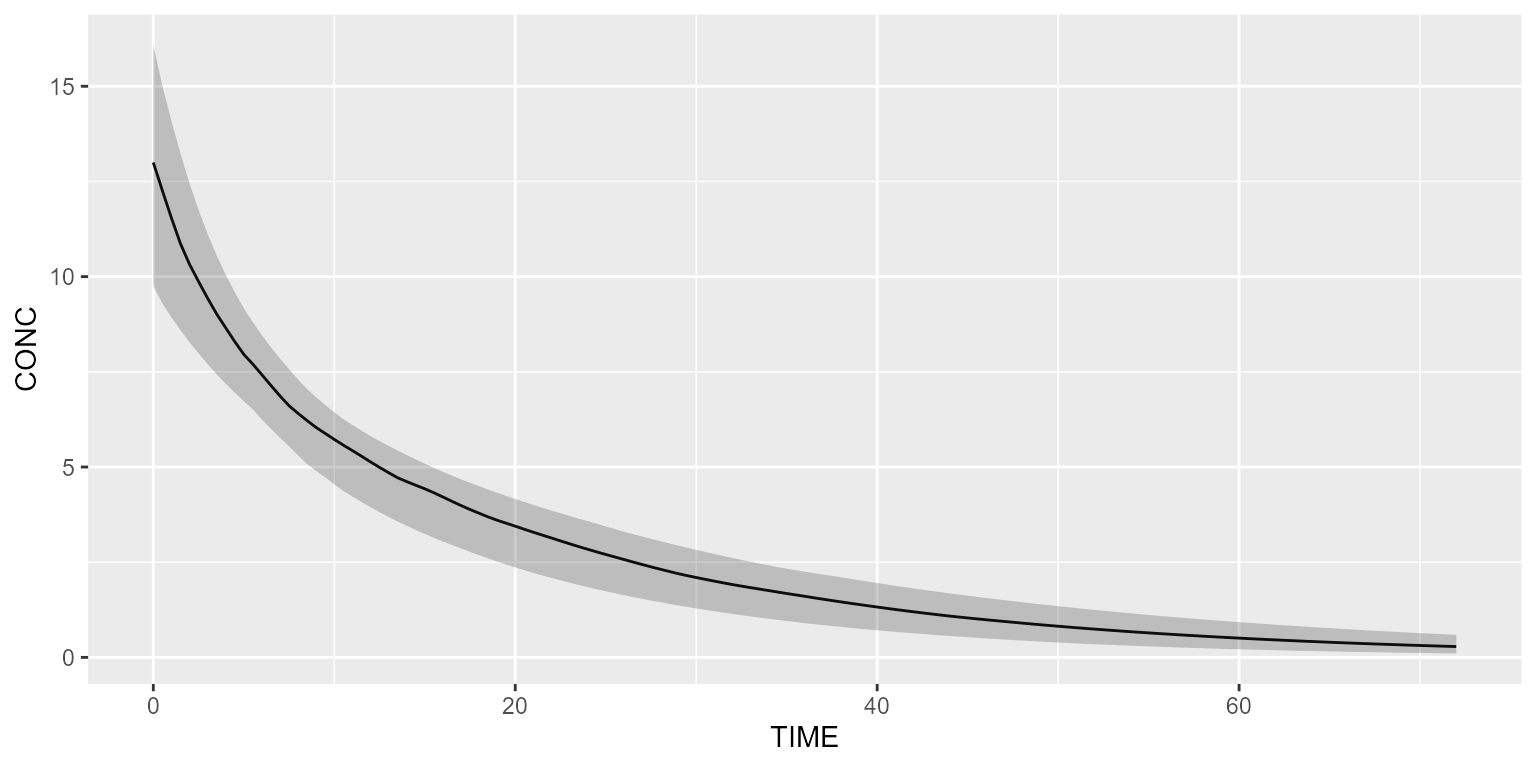
## A_OUTPUT (CMT=3)Let’s now create a dataset with observations-only.
ds <- Dataset(50) %>%
add(Observations(times=seq(0,72, by=0.5)))We can now simulate this model:
results <- model %>% simulate(dataset=ds, seed=1)
shadedPlot(results, "CONC")