There are 2 ways to implement bioavailability in CAMPSIS:
- in the model: bioavailability is defined for each compartment
- in the dataset: bioavailability is defined for each bolus or infusion
In the first case, the simulation engine will take care of the bioavailability. In the second case, CAMPSIS will adapt automatically the amount injected through the dataset (AMT column).
Bioavailability implemented in model
Let’s use a 2-compartment model with absorption compartment to illustrate how this can be achieved.
model <- model_suite$nonmem$advan4_trans4For this example, we’re going to define a bioavailability
F1 for this absorption compartment.
First let’s create a new parameter F1, log-normally
distributed with a median of 0.75 and 10% CV.
model <- model %>% add(Theta(name="F1", value=0.75))
model <- model %>% add(Omega(name="F1", value=10, type="cv%"))Now, let’s add an equation to the drug model to define
F1.
Finally, we need to tell CAMPSIS that F1 corresponds to
a bioavailability.
model <- model %>% add(Bioavailability(compartment=1, rhs="F1"))Our persisted drug model would look like this:
model## [MAIN]
## KA=THETA_KA*exp(ETA_KA)
## CL=THETA_CL*exp(ETA_CL)
## V2=THETA_V2*exp(ETA_V2)
## V3=THETA_V3*exp(ETA_V3)
## Q=THETA_Q*exp(ETA_Q)
## S2=V2
## F1=THETA_F1*exp(ETA_F1)
##
## [ODE]
## d/dt(A_DEPOT)=-KA*A_DEPOT
## d/dt(A_CENTRAL)=KA*A_DEPOT + Q*A_PERIPHERAL/V3 + (-CL/V2 - Q/V2)*A_CENTRAL
## d/dt(A_PERIPHERAL)=-Q*A_PERIPHERAL/V3 + Q*A_CENTRAL/V2
## d/dt(A_OUTPUT)=CL*A_CENTRAL/V2
## F=A_CENTRAL/S2
##
## [F]
## A_DEPOT=F1
##
## [ERROR]
## CONC=F
## CONC_ERR=CONC*(EPS_PROP + 1)
##
##
## THETA's:
## name index value fix
## 1 KA 1 1.00 FALSE
## 2 CL 2 5.00 FALSE
## 3 V2 3 80.00 FALSE
## 4 V3 4 20.00 FALSE
## 5 Q 5 4.00 FALSE
## 6 F1 6 0.75 FALSE
## OMEGA's:
## name index index2 value fix type
## 1 KA 1 1 0.025 FALSE var
## 2 CL 2 2 0.025 FALSE var
## 3 V2 3 3 0.025 FALSE var
## 4 V3 4 4 0.025 FALSE var
## 5 Q 5 5 0.025 FALSE var
## 6 F1 6 6 10.000 FALSE cv%
## SIGMA's:
## name index index2 value fix type
## 1 PROP 1 1 0.025 FALSE var
## No variance-covariance matrix
##
## Compartments:
## A_DEPOT (CMT=1)
## A_CENTRAL (CMT=2)
## A_PERIPHERAL (CMT=3)
## A_OUTPUT (CMT=4)Now, let’s now give a simple bolus and simulate with and without
F1.
ds1 <- Dataset(50) %>%
add(Bolus(time=0, amount=1000)) %>%
add(Observations(times=seq(0,24,by=0.5)))
results_f1 <- model %>% simulate(dataset=ds1, seed=1)
results_no_f1 <- model_suite$nonmem$advan4_trans4 %>% simulate(dataset=ds1, seed=1)
gridExtra::grid.arrange(shadedPlot(results_f1, "CONC"), shadedPlot(results_no_f1, "CONC"), nrow=1)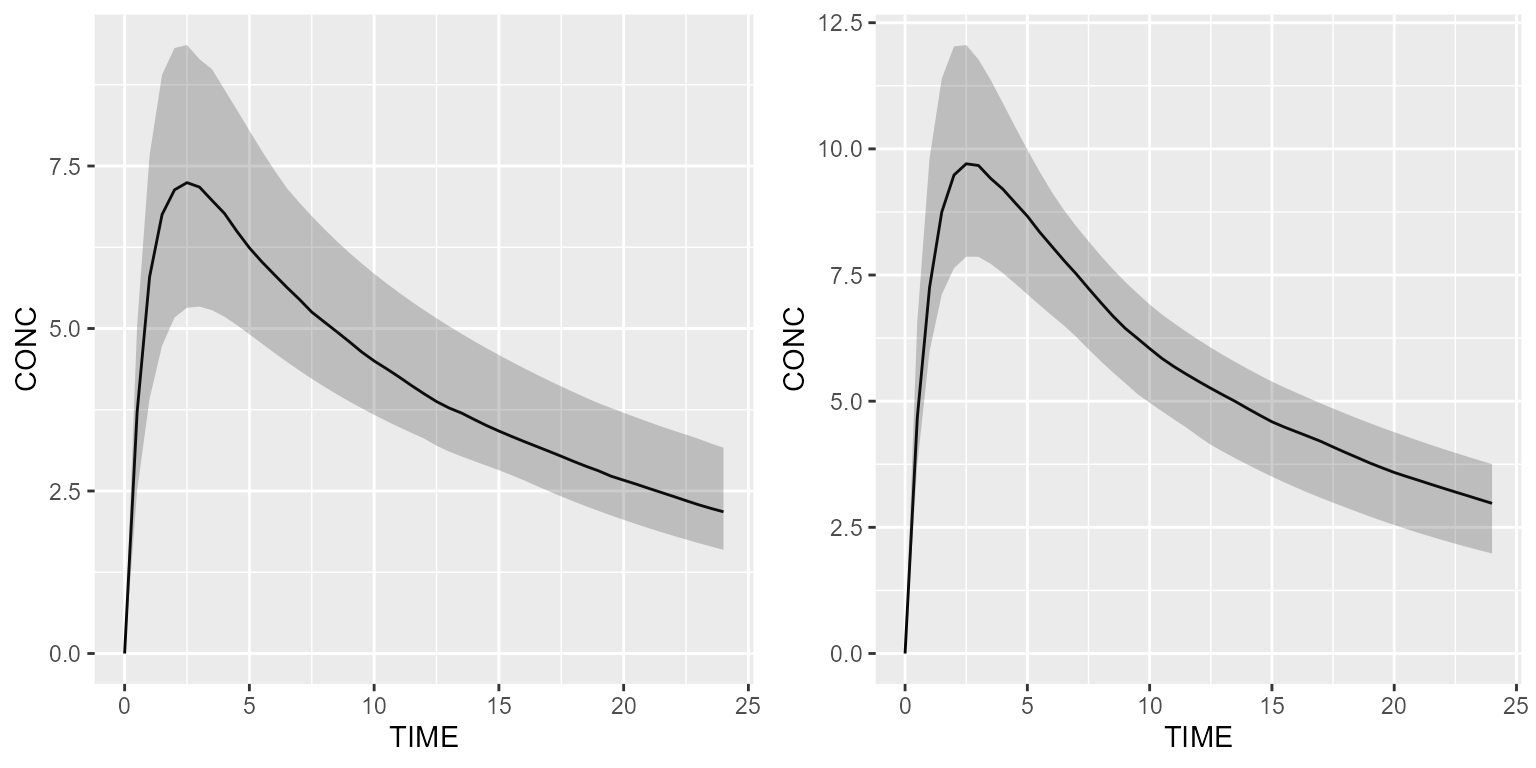
Bioavailability implemented in dataset
The same simulation can be performed by adapting the column
AMT in the dataset.
First, we need to sample F1 values. This can be done as
follows:
distribution <- ParameterDistribution(model=model, theta="F1", omega="F1") %>%
sample(50L)We can then pass the pre-sampled distribution.
ds2 <- Dataset(50) %>%
add(Bolus(time=0, amount=1000, f=distribution)) %>%
add(Observations(times=seq(0,24,by=0.5)))Let’s have a look at the dataset, in its table form, and if we look at the doses only:
## # A tibble: 6 × 9
## ID ARM TIME EVID MDV AMT CMT RATE DOSENO
## <dbl> <int> <dbl> <int> <int> <dbl> <chr> <dbl> <int>
## 1 1 0 0 1 1 705. 1 0 1
## 2 2 0 0 1 1 764. 1 0 1
## 3 3 0 0 1 1 690. 1 0 1
## 4 4 0 0 1 1 879. 1 0 1
## 5 5 0 0 1 1 775. 1 0 1
## 6 6 0 0 1 1 691. 1 0 1Finally, we can simulate the original model using this new dataset.
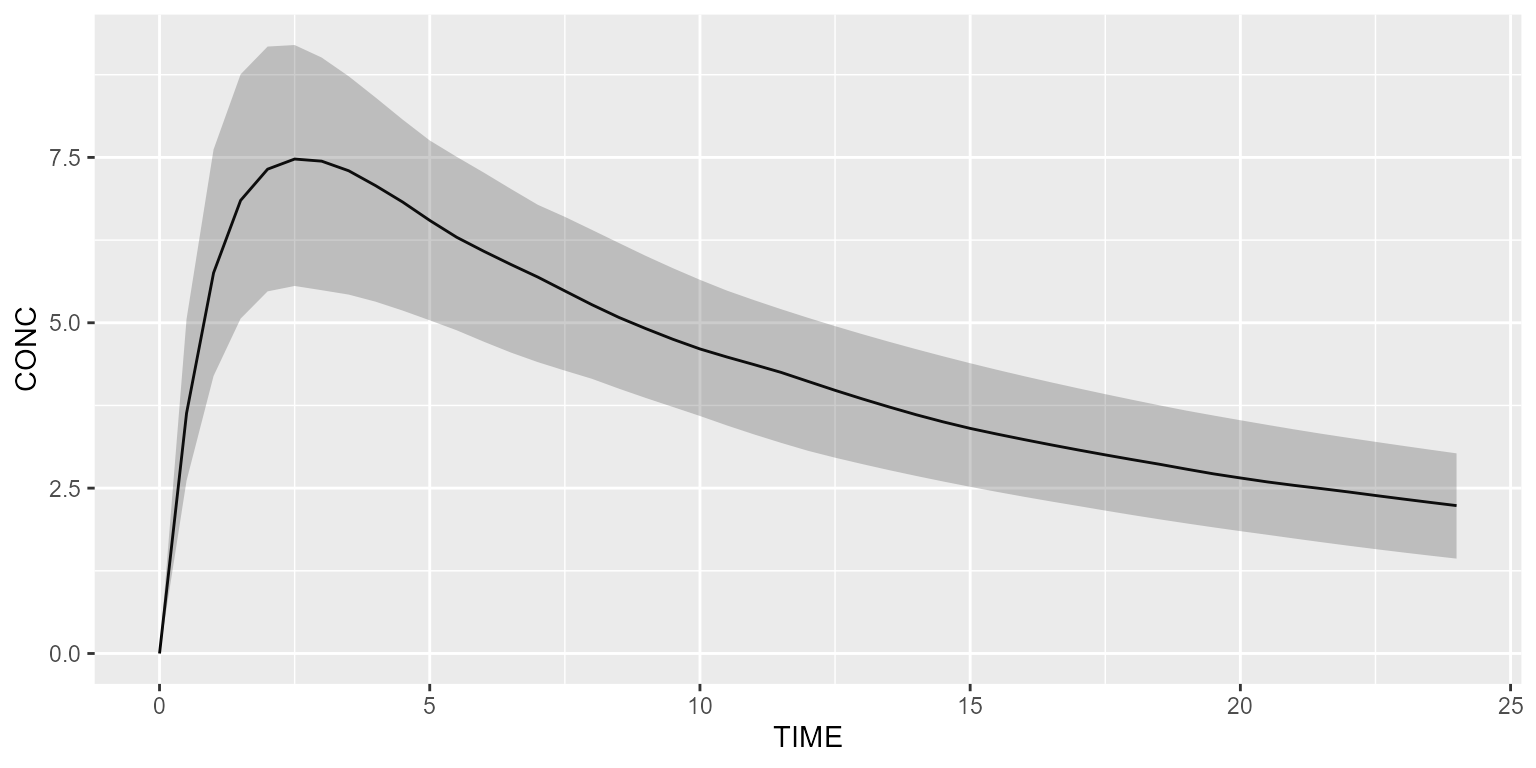
results_f1 <- model_suite$nonmem$advan4_trans4 %>% simulate(dataset=ds2, seed=1)
shadedPlot(results_f1, "CONC")