This vignette shows how a simulation can be replicated.
Simulate uncertainty on percentiles
Assume the following model is used. This model is a 2-compartment model without absorption compartment which has been fitted on some data.
model <- model_suite$testing$other$my_model1It contains a variance-covariance matrix with the uncertainty on all the estimated parameters.
model## [MAIN]
## CL=THETA_CL*exp(ETA_CL)
## V1=THETA_V1*exp(ETA_V1)
## V2=THETA_V2
## Q=THETA_Q
## S1=V1
##
## [ODE]
## d/dt(A_CENTRAL)=Q*A_PERIPHERAL/V2 + (-CL/V1 - Q/V1)*A_CENTRAL
## d/dt(A_PERIPHERAL)=-Q*A_PERIPHERAL/V2 + Q*A_CENTRAL/V1
## d/dt(A_OUTPUT)=CL*A_CENTRAL/V1
## F=A_CENTRAL/S1
##
## [DURATION]
## A_CENTRAL=5
##
## [ERROR]
## CP=F
## OBS_CP=CP*(EPS_PROP + 1)
## Y=OBS_CP
##
##
## THETA's:
## name index value fix se rse%
## 1 CL 1 4.76756 FALSE 0.1163899 2.441288
## 2 V1 2 82.64090 FALSE 1.9256999 2.330202
## 3 V2 3 19.53960 FALSE 1.5382328 7.872386
## 4 Q 4 3.81451 FALSE 0.4726151 12.389929
## OMEGA's:
## name index index2 value fix type se rse%
## 1 CL 1 1 0.0222955 FALSE var 0.004867504 21.83178
## 2 V1 2 2 0.0182225 FALSE var 0.005172881 28.38733
## SIGMA's:
## name index index2 value fix type se rse%
## 1 PROP 1 1 0.0244587 FALSE var 0.0008126574 3.32257
## Variance-covariance matrix available (see ?getVarCov)
##
## Compartments:
## A_CENTRAL (CMT=1)
## A_PERIPHERAL (CMT=2)
## A_OUTPUT (CMT=3)We are interested to see the uncertainty on the simulated concentration percentiles over time. Let’s mimic the protocol that was implemented in the study.
ds <- Dataset(50) %>%
add(Infusion(time=(0:6)*24, amount=1000, compartment=1)) %>%
add(Observations(times=seq(0, 7*24)))Let’s now simulate this model with parameter uncertainty.
Argument replicates specifies how many times the simulation
is replicated.
Argument outfun is a function that is going to be called
after each simulation on the output data frame.
results <- model %>% simulate(dataset=ds, replicates=10, outfun=~PI(.x, output="Y"), seed=1)
results %>% head()## # A tibble: 6 × 4
## # Groups: TIME [2]
## replicate TIME metric value
## <int> <dbl> <chr> <dbl>
## 1 1 0 med 0
## 2 1 0 low 0
## 3 1 0 up 0
## 4 1 1 med 2.21
## 5 1 1 low 1.51
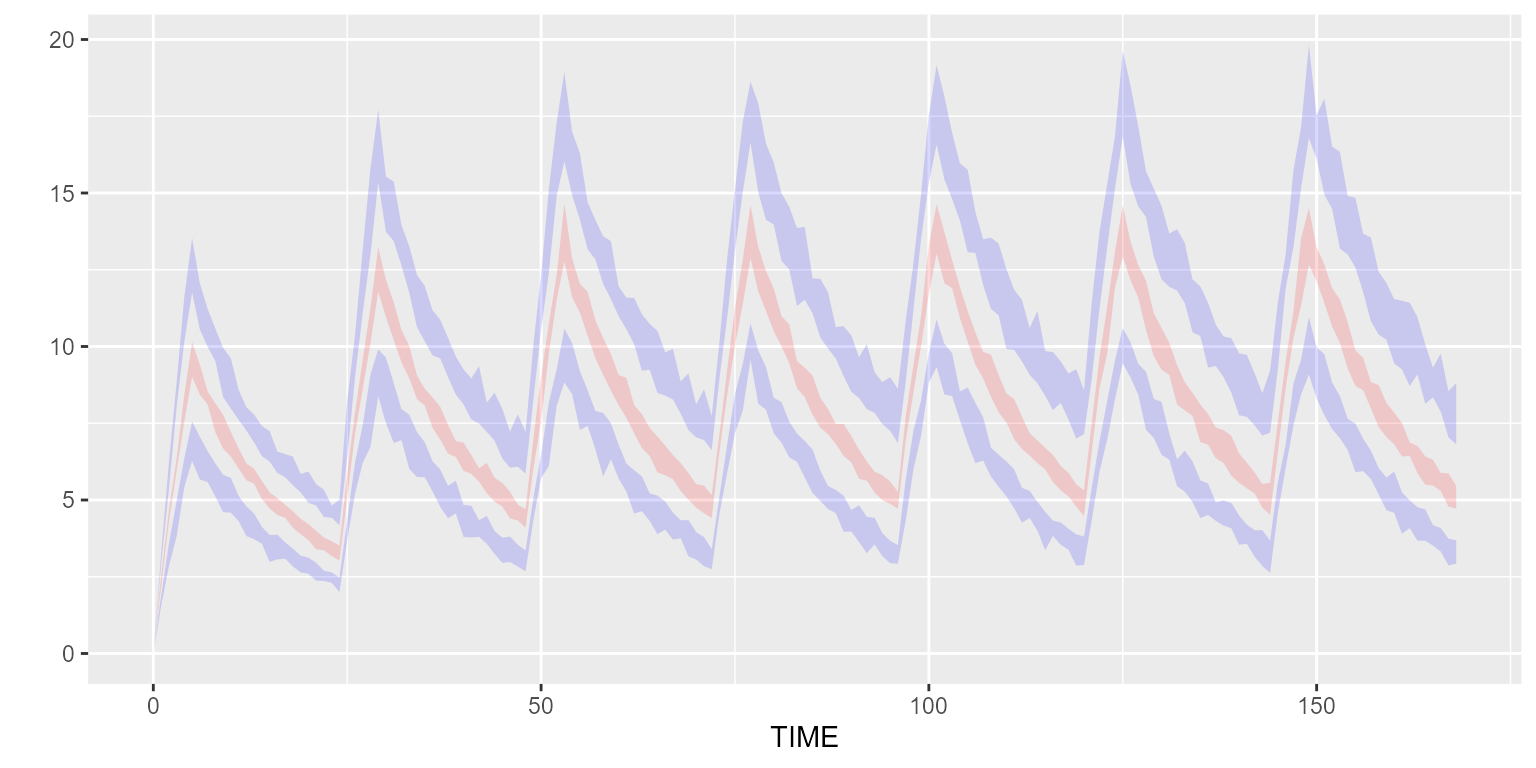
## 6 1 1 up 3.15Function vpcPlot allows to quickly visualize such
results.
vpcPlot(results)